Sunday, October 30, 2022
Wednesday, October 26, 2022
Tuesday, October 25, 2022
Daftar Tugas Pengelola Laboratorium Di Sekolah
Tips ArtikelKaca Arloji: Pengertian, Fungsi, Penggunaan Dan Cara Perawatannya
Tips ArtikelJual Constant Temperature Drying Oven
Tips ArtikelPengertian Dan Fungsi Corong Pemisah (Separator Funnel)
Tips ArtikelTabung Reaksi: Pengertian, Fungsi Dan Cara Perawatannya
Tips ArtikelTuesday, October 4, 2022
Cara Perawatan Inkubator Laboratorium
Sunday, November 22, 2020
An Evaluation of Backpropagation Interpretability for Graph Classification with Deep Learning
A summary of the IEEE BigData 2020 research paper by Kenneth Teo Tian Shun, Eko Edita Limanta, and Arijit Khan
[Graph Classification] Graph data are ubiquitous in many domains, such as social networks, knowledge graphs, biological networks, geo-spatial road networks, and internet-of-things. There are plenty of interest and applications in developing high-quality machine learning techniques for classifying graph objects, including cheminformatics (e.g., compounds that are active or inactive for some target) [1], [2] and bioinformatics (e.g., classifying proteins into different families) [3], malware detection and classification with call graphs [18]. There are two research directions for classification tasks on graphs: (1) Given a single large graph, infer labels of individual nodes (the node classification problem) [4], [5]; and (2) given a set of graphs with different structure and sizes, predict the class labels of unseen graphs (the graph classification problem) [6], [7], [19]. We focus on the second problem — graph classification, which is more difficult because graph datasets contain graphs with varying numbers of nodes and edges.
[Why Deep Learning in Graph Classification] Unlike images, general graphs do not have regular grid-like structures, and the neighborhood size of each node varies to a great extent. The lack of ordered vector representation limits the applicability of machine learning on graphs, and consequently, makes it difficult to build a classifier directly over the graph space [19], [7]. In the past, graph kernels (e.g., random walks, shortest paths, cyclic patterns, subtrees, graphlets, and subgraph kernels) [8] and feature based classification (e.g., frequent and significant subgraphs) methods [9], [10] were developed. Due to the challenges of feature engineering, coupled with the hardness of subgraph mining and isomorphism testing, deep learning methods are increasingly becoming popular. In particular, the end-to-end nature of learning in convolutional neural networks (CNNs) and their ability to extract localized spatial features, have turned them into powerful tools for learning from a large corpus of data including graphs. We focus on three state-of-the-art graph convolutional neural networks (GCNNs): GCNN with global average pooling (GCNN+GAP)[11], [5], DGCNN [19], and DIFFPOOL [7], as well as their variants.
[Interpretability Challenges in Deep Learning-based Graph Classification] Deep learning models and neural networks are “black-box” — it is inherently difficult to understand which aspects of the input data drive the decisions of the network. Interpretability can improve the model’s transparency related to fairness, privacy, and other safety challenges, thus enhancing the trust in decision-critical applications [12], [13]. Network data are complex, noisy, and content-rich. End users (e.g., data scientists, business analysts) often find them difficult to query and explore [14]. Hence, it is even more critical to provide human-understandable interpretation over classification results on such datasets. While interpretability for GCNNs is an open area of research, inspired by recent interpretability tools for CNNs, in this work we investigate three backpropagation based post-hoc interpretability methods: (1) saliency map with contrastive gradients (CG) [15], (2) gradient-weighted class activation mapping (Grad-CAM) [16], and (3) deep learning important features (DeepLIFT) [17]. We conduct a thorough experimental evaluation of these interpretability methods in conjunction with three state-of-the-art graph convolutional neural networks: GCNN+GAP [11], [5], DGCNN [19], and DIFFPOOL [7], and with their variants.
We design novel strategies for integrating them, measuring their efficiency and performance both qualitatively and quantitatively. We implement GCNNs and interpretability tools in a common environment, using same hyperparameters selection criteria, and present extensive empirical comparisons over five real-world graph datasets from different categories. We report their quantitative, visualization, and active subgraph based performance, compare them with the classic significant subgraph mining results [10], summarize their trade-offs, and provide guidelines for researchers and practitioners.
[Summary and our recommendation] The following table summarizes our recommendation levels of each interpretability method according to different performance metrics. The scale is from 1 to 4 stars, larger number of stars stands for higher recommendation. Clearly, there is no single winner.
(1) Considering various trade-offs, our recommended interpretability tools for graph classification are DeepLIFT, GradCAM, in combination with recent (and more accurate) GCNNs: DGCNN, DIFFPOOL, and its variant DIFFPOOL_D. (2) Grad-CAM is useful for class-discriminative visualization, whereas DeepLIFT can identify differences between two individual instances. (3) Finally, DIFFPOOL and DIFFPOOL_D can assist visualizing node clustering at various granularity, and this has the potential to identify active subgraphs, e.g., active functional groups for biological networks classification.
We open sourced an end-to-end framework (https://github.com/tsKenneth/interpretable-graph-classification) in which one can plug and play with different graph datasets, GCNNs, and backpropagation-based interpretability tools. In future, it would be interesting to test and model how high accuracy and fidelity could impact the user’s trust in the underlying graph classification tool, as well as to conduct usability analysis due to higher contrastivity, sparsity, explanation subgraph size, and efficiency.
For more information, click here for our paper. Code: GitHub.
References
[1] Z. Wu, B. Ramsundar, E. N. Feinberg, J. Gomes, C. Geniesse, A. S. Pappu, K. Leswing, and V. S. Pande, “MoleculeNet: A Benchmark for Molecular Machine Learning”, CoRR, vol. abs/1703.00564, 2017.
[2] D. Duvenaud, D. Maclaurin, J. A.-Iparraguirre, R. G.-Bombarelli, T. Hirzel, A. A.-Guzik, and R. P. Adams, “Convolutional Networks on Graphs for Learning Molecular Fingerprints,” NeurIPS, 2015.
[3] R. Sharan, S. Suthram, R. M. Kelley, T. Kuhn, S. McCuine, P. Uetz, T. Sittler, R. M. Karp, and T. Ideker, “Conserved Patterns of Protein Interaction in Multiple Species”, PNAS, vol. 102, no. 6, 2005.
[4] J. Bruna, W. Zaremba, A. Szlam, and Y. LeCun, “Spectral Networks and Locally Connected Networks on Graphs,” ICLR, 2014.
[5] T. N. Kipf and M. Welling, “Semi-Supervised Classification with Graph Convolutional Networks”, ICLR, 2017.
[6] D. Duvenaud, D. Maclaurin, J. A.-Iparraguirre, R. G.-Bombarelli, T. Hirzel, A. A.-Guzik, and R. P. Adams, “Convolutional Networks on Graphs for Learning Molecular Fingerprints,” NeurIPS, 2015.
[7] Z. Ying, J. You, C. Morris, X. Ren, W. L. Hamilton, and J. Leskovec, “Hierarchical Graph Representation Learning with Differentiable Pooling”, NeurIPS, 2018.
[8] S. V. N. Vishwanathan, N. N. Schraudolph, R. Kondor, and K. M. Borgwardt, “Graph Kernels”, J. Mach. Learn. Res., vol. 11, 2010.
[9] X. Yan, H. Cheng, J. Han, and P. S. Yu, “Mining Significant Graph Patterns by Leap Search”, SIGMOD, 2008.
[10] S. Ranu and A. K. Singh, “GraphSig: A Scalable Approach to Mining Significant Subgraphs in Large Graph Databases”, ICDE, 2009.
[11] M. Lin, Q. Chen, and S. Yan, “Network In Network”, ICLR, 2014.
[12] M. Du, N. Liu, and X. Hu, “Techniques for Interpretable Machine Learning”, Commun. ACM, vol. 63, no. 1, 2020.
[13] Z. C. Lipton, “The Mythos of Model Interpretability”, Commun. ACM, vol. 61, no. 10, 2018.
[14] N. Jayaram, A. Khan, C. Li, X. Yan, and R. Elmasri, “Querying Knowledge Graphs by Example Entity Tuples”, IEEE Trans. Knowl. Data Eng., vol. 27, no. 10, 2015.
[15] K. Simonyan, A. Vedaldi, and A. Zisserman, “Deep Inside Convolutional Networks: Visualising Image Classification Models and Saliency Maps”, ICLR, 2013.
[16] R. R. Selvaraju, M. Cogswell, A. Das, R. Vedantam, D. Parikh, and D. Batra, “Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization”, ICCV, 2017.
[17] A. Shrikumar, P. Greenside, and A. Kundaje, “Learning Important Features Through Propagating Activation Differences”, ICML, 2017.
[18] M. Fan, X. Luo, J. Liu, M. Wang, C. Nong, Q. Zheng, and T. Liu, “Graph Embedding based Familial Analysis of Android Malware using Unsupervised Learning,” ICSE, 2019.
[19] M. Zhang, Z. Cui, M. Neumann, and Y. Chen, “An End-to-End Deep Learning Architecture for Graph Classification”, AAAI, 2018.
Friday, June 5, 2020
Mining Top-k Pairs of Correlated Subgraphs in a Large Network
[Background and Problem] A large body of work exists on mining recurring structural patterns among a group of nodes in the form of frequent subgraphs [1, 2]. However, can we mine recurring patterns among the frequent subgraphs themselves? In this paper, we explored this question by mining correlated pairs of frequent subgraphs. Correlated subgraphs are different from frequent subgraphs due to the flexibility in connections between constituent subgraph instances. To elaborate, in Figure 1, we highlight three regions inside the chemical structure of Taxol, an anti-cancer drug, where CCCH and O occur closely albeit connected in different ways in all three instances. For simplicity, we do not consider the edge types (i.e., single or double bonds) in this example. This figure illustrates that while CCCH and O form a frequently occurring correlated pair of subgraphs, the individual instances (for example, HCC(-O)C) may not be frequent patterns themselves. Therefore, existing frequent subgraphs mining techniques cannot mine such pairs of correlated subgraph patterns.
[Application Scenarios] Correlated subgraphs mining (CSM) will be useful for many applications. For example, it can be used in the identification of co-operative functions in biological networks. In a genome graph of an individual, a node represents a gene and each edge denotes the interaction between two genes. In practice, there are some combinations of dominant genes that co-occur frequently, and these are more likely to express critical phenotypes of the individual. Previous studies show that some pairs of dominant gene combinations co-occur in each individual, and such co-occurring patterns often reflect the joint functionality that is needed for co-operative biochemical functioning such as chemical bonding and binding sites interactions [18]. Based on pairs of correlated genes, we can predict co-operative biological functions of an individual.
To demonstrate the utility of CSM, we present insights derived from the correlated pairs mined in the following datasets.
Figure 2: (a-d) Correlated subgraph pairs <Q1, Q2> and <Q3, Q4> in Yeast. (e) Correlated pair in LastFM. CP denotes Coldplay and RH denotes Radiohead. (f) Correlated pair in Coauthor.
LastFM [20]. Figure 2e depicts a correlated pair from the top-k set. This pair showcases that communities of users who like the music band Coldplay, are often in close proximity to communities of users who like Radiohead. These two bands are contemporaries, originate from UK and they are often compared on social media.
Coauthor [21]. Figure 2f presents a pair from the top-10 list in the Coauthor (DBLP) network. Nodes correspond to authors and node labels denote the most frequent publication venue of the authors. In this regard, the presented pair is interesting since it reveals proximity between the networking community (ICCCN) and the architecture community (SIGARCH). While most of the other pairs are between conferences centered on similar areas, this pair shows that there is high collaboration between networking and architecture communities. More importantly, this pair reveals that correlated subgraphs may unearth non-trivial connections between communities.
[Challenges and Differences with Related Work]
[Our Algorithmic Contributions] In spite of the aforementioned challenges and fundamental differences with existing literature, it is still desirable to obtain CSM results within minutes over real networks containing millions of nodes. In this work, we achieve this task with our novel algorithmic contributions.
Sunday, April 19, 2020
Reliability Maximization in Uncertain Graphs
Reliability Maximization in Uncertain Graphs
[Our Problem] Given an uncertain graph, a source node s, a target node t, a probability threshold ζ ∈ (0, 1], and a small positive integer k, we investigate to find the top-k edges to add in the graph, each with probability ζ, so that the reliability from the source s to the target t is maximized.
[Application Scenarios] (1) In mobile ad-hoc networks, the connectivity between sensor nodes and devices is estimated using noisy measurements, thus leading to edges naturally associated with a probability of existence [7]. Road networks can be modeled as uncertain graphs because of unexpected traffic congestion [6]. In these networks, creating new connections between nodes (e.g., building new roads, flyovers, adding Ethernet cables) is limited by physical constraints and budget. One can introduce only k new edges where k is decided based on resource constraints. (2) In social networks, finding k best shortcut edges could maximize the information diffusion probability from an early adopter to a target customer [8], thus the network host can actively recommend these links to the respective users. (3) In case of protein-interaction networks, interactions are established for a limited number of proteins through noisy and error-prone experiments—each edge is associated with a probability accounting for the existence of the interaction. Therefore, finding the top-k shortcut edges can assist in de-noising protein-interaction networks [9].
[Algorithmic Contributions] In spite of such challenges, we devise efficient algorithms. (1) We investigate several baselines, analyse their complexities, and discuss the shortcomings. They include individual top-k edge selection, hill-climbing method, centrality-based method, and eigenvalue-based method. (2) We eliminate the search space (can be up to O(n^2) in sparse real networks) via finding the promising nodes with high original reliability from the source, or to the target. (3) After adding all possible missing edges only between pairs of promising nodes, we further filter out those paths with low probabilities from s to t. The intuition is that what really matters in computing the reliability between two nodes is the set of highly reliable paths between them [10]-[12]. (4) We adapt the single-path-inclusion algorithm in [10] to our problem setting, and refine it in a path-batch manner to significantly improve the accuracy.
[Other Queries] We also consider a restricted and one extended version of our problem to support a wider family of queries. (1) In the restricted version, the reliability is estimated only by the most reliable path, thus it can be solved in polynomial-time. (2) In the extended version, multiple sources and targets can be provided as input. We investigate several possible reliability aggregation functions. The proposed algorithms are generalized to each case.
[Case Study] The Intel Lab Data (http://db.csail.mit.edu/labdata/labdata.html) dataset contains the sensor network information with 54 sensors deployed in the Intel Berkeley Research Lab (map given in Figures 2 and 3). The probabilities on links denote the percentages of messages from a sender successfully reached to a receiver. Only those links with probabilities higher than the average value (0.33) are shown in the figures, and the thickness represents link probabilities. Due to budget constraints, only 3 new links are allowed for each case. We further assume that the probability of each new link would be the same as the average edge probability of the original dataset, that is, 0.33. We notice that if two sensors are more than 20 meters away, the original link probability between them is usually close to 0. Thus, we only allow establishing new links between a pair of sensors that are at most 15 meters away.
For case (1), sensor 46 has very weak connections from outside, while sensor 21 is connected with the bottom part of the lab, where a very dense network exists. Therefore, our solution for this case is to connect sensor 46 with sensors in the bottom part of the lab. By establishing three links 2 to 46, 35 to 46, and 37 to 46, we improve the reliability between 21 to 46 from 0.40 to 0.88.
For case (2), notice in Figure 7 that the sensors in the center part of lab are well-connected, and the links in this region are thicker than those in the bottom part. The source sensor 15 has a few connections with sensors in the bottom part, but no link with those in the center part. The destination sensor 40 has limited connections beyond its physical neighbors. Existing configuration offers a poor reliability of 0.28 for the connection between source and destination which we like to improve. The smart decision made by our algorithm is as follows: First, connect sensor 35 to 40, thus making sensor 35 a bridge between the center and the bottom region of the network; Second, enable connection from sensor 15 to the center part (by establishing link from 15 to 10, and from 15 to 11). This results in 0.58 overall reliability from sensor 15 to 40, which is more than double of the original reliability value.
These results illustrate how our proposed solution for the budgeted reliability maximization problem can be useful in solving real-life problems.
[3] L. G. Valiant, “The Complexity of Enumeration and Reliability Problems”, SIAM J. on Computing, vol. 8, no. 3, pp. 410–421, 1979.
[4] K. K. Aggarwal, K. B. Misra, and J. S. Gupta, “Reliability Evaluation: A Comparative Study of Different Techniques” Micro. Rel., vol. 14, no. 1, 1975.
[5] D. Kempe, J. M. Kleinberg, and E. Tardos, “Maximizing the Spread of Influence through a Social Network”, in KDD, 2003.
[6] M. Hua and J. Pei, “Probabilistic Path Queries in Road Networks: Traffic Uncertainty aware Path Selection”, in EDBT, 2010.
[7] J. Ghosh, H. Q. Ngo, S. Yoon, and C. Qiao, “On a Routing Problem Within Probabilistic Graphs and its Application to Intermittently Connected Networks”, in INFOCOM, 2007.
[8] C. Chen, H. Tong, B. A. Prakash, T. Eliassi-Rad, M. Faloutsos, and C. Faloutsos, “Eigen-Optimization on Large Graphs by Edge Manipulation”, ACM Trans. Knowl. Discov. Data, vol. 10, no. 4, pp. 49:1–49:30, 2016.
[9] O. Kuchaiev, M. Rasajski, D. J. Higham, and N. Przulj, “Geometric De-noising of Protein-Protein Interaction Networks”, PLOS Computational Biology, vol. 5, no. 8, pp. 1–10, 08 2009.
[10] A. Khan, F. Bonchi, F. Gullo, and A. Nufer, “Conditional Reliability in Uncertain Graphs”, IEEE Trans. Knowl. Data Eng., vol. 30, no. 1, pp. 2078–2092, 2018.
[11] W. Chen, C.Wang, and Y.Wang, “Scalable Influence Maximization for Prevalent Viral Marketing in Large-Scale Social Networks”, in KDD, 2010.
[12] A. Khan, B. Zehnder, and D. Kossmann, “Revenue Maximization by Viral Marketing: A Social Network Host’s Perspective”, in ICDE, 2016.
Tuesday, March 17, 2020
Semantic Guided and Response Times Bounded Top-k Similarity Search over Knowledge Graphs
Background: Knowledge graphs (such as DBpedia [1], Yago [2], and Freebase [3]) have been constructed in recent years, managing large-scale and real-world facts as a graph [4]. In such graphs, each node represents an entity with attributes, and each edge denotes a relationship between two entities.
Fig. 1: An example of structural mismatch with query graphs: different users may employ different query graphs (top) to find all cars made in Germany. Four correct graph matches in DBpedia are provided (bottom). Only G2Q can retrieve partial correct answers having the same schemas as 1 , because the 1-hop edge assembly cannot match any n-hop (n > 1) paths.
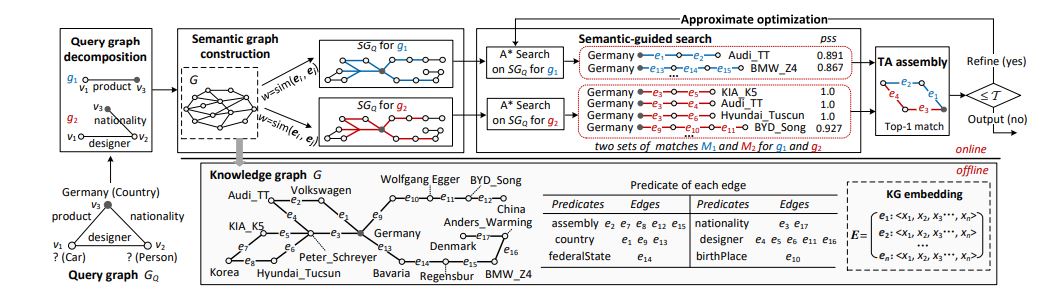
Fig. 2: A running example of our approach, including offline KG embedding (bottom right) and online query processing. Four components of the online part are: (1) Query graph decomposition, (2) Semantic graph construction, (3) Semantic-guided search, and (4) Threshold Algorithm-based assembly. An approximate optimization is applied for response-time-bounded query. All predicates in the example knowledge graph are provided in a table (bottom middle).
[1] P. N. Mendes, M. Jakob, and C. Bizer, “Dbpedia: A multilingual crossdomain knowledge base.” in LREC, 2012, pp. 1813–1817.
[2] J. Hoffart, F. M. Suchanek, K. Berberich, and G. Weikum, “Yago2: A spatially and temporally enhanced knowledge base from wikipedia,” Artificial Intelligence, vol. 194, pp. 28–61, 2013.
[3] K. Bollacker, R. Cook, and P. Tufts, “Freebase: A shared database of structured general human knowledge,” in AAAI, 2007, pp. 1962–1963.
[4] X. Huang, J. Zhang, D. Li, and P. Li, “Knowledge graph embedding based question answering,” in WSDM, 2019, pp. 105–113.
[5] R. V. Guha, R. McCool, and E. Miller, “Semantic search,” in WWW, 2003, pp. 700–709.
[6] A. Khan, Y. Wu, C. C. Aggarwal, and X. Yan, “Nema: Fast graph search with label similarity,” PVLDB, vol. 6, no. 3, pp. 181–192, 2013.
[7] L. Zou, R. Huang, H. Wang, J. X. Yu, W. He, and D. Zhao, “Natural language question answering over rdf: a graph data driven approach,” in SIGMOD, 2014, pp. 313–324.
[8] S. Yang, Y. Wu, H. Sun, and X. Yan, “Schemaless and structureless graph querying,” PVLDB, vol. 7, no. 7, pp. 565–576, 2014.
[9] S. Yang, F. Han, Y. Wu, and X. Yan, “Fast top-k search in knowledge graphs,” in ICDE, 2016, pp. 990–1001.
[10] J. Jin, S. Khemarat, and L. Gao, “Querying web scale information networks via bounding matching scores,” in WWW, 2015, pp. 527–537.
[11] W. Zheng, L. Zou, X. Lian, J. X. Yu, S. Song, and D. Zhao, “How to build templates for rdf question/answering: An uncertain graph similarity join approach,” in SIGMOD, 2015, pp. 1809–1824.
[12] S. Han, L. Zou, J. X. Yu, and D. Zhao, “Keyword search on rdf graphs-a query graph assembly approach,” in CIKM, 2017, pp. 227–236.
[13] S. Shekarpour, E. Marx, S. Auer, and A. Sheth, “Rquery: rewriting natural language queries on knowledge graphs to alleviate the vocabulary mismatch problem,” in AAAI, 2017, pp. 3936–3943.
[14] W. Zheng, L. Zou, W. Peng, X. Yan, S. Song, and D. Zhao, “Semantic sparql similarity search over rdf knowledge graphs,” PVLDB, vol. 9, no. 11, pp. 840–851, 2016.
[15] “Qald-4,” http://qald.aksw.org/index.php?x=challenge&q=4.
[16] S. S. Bhowmick, B. Choi, and S. Zhou, “Vogue: Towards a visual interaction-aware graph query processing framework,” in CIDR, 2013.
[17] R. Fagin, A. Lotem, and M. Naor, “Optimal aggregation algorithms for middleware,” in PODS, 2001.
[18] A. Bordes, N. Usunier, A. Garc´ıa-Duran, J. Weston, and O. Yakhnenko, “Translating embeddings for modeling multi-relational data,” in NIPS, 2013, pp. 2787–2795.